Research
Our lab has three main interests:
First, mechanistic, to reveal the missing components that allow the transport of signaling molecules. It initiated when we identified the first gibberellin transporter in plants (Nature Comm, 2016) and continued with novel transport mechanisms explaining how hormones translocate from the site of synthesis to their active site: auxin (Nature Comm 2018, Nature Comm 2021, EMBO Reports 2023), cytokinin (Nature Plants 2023a), gibberellin (Nature Plants 2023b), and ABA (Science Advances 2021).
Second, How do plants sense and respond to water stress? Centered around a $10M ERC-SyG grant we are leading with Malcolm Bennett (Nottingham University), Christine Ziegler (University of Regensburg) and Thorsten Hamann (NTNU), that initiated in 2024 (Link), we show that Hydrosignals movement is required for the plant’s response to water deficiency (Mehra et al., Science 2022). More details soon.
Third, developing next-generation genetics tools/approaches. To reveal the missing genetic mechanisms, our lab is developing novel genome-scale genetic tools that allow for tackling technological barriers in plant sciences. These tools primarily focus on overcoming functional redundancy at large scales (Nature Plants 2023, and unpublished). The CRISPR-based library tools we are developing are transforming how plant genetics is carried out and are currently being adopted by seed companies to enhance breeding programs.



More Details
Developing next-generation multi-targeted CRISPR genetic toolboxes in plants
Functional redundancy is a significant obstacle in molecular genetics and breeding, considerably limiting our ability to improve plant health and food production. Our lab developed and validated a genome-scale CRISPR toolbox that overcomes functional redundancy in Arabidopsis by simultaneously targeting multiple gene-family members (Nature Plants 2023).
The Multi-Knock tool, which reveals genetically hidden components on large scales, is already changing plant genetics and is currently being used in collaboration with multiple institutes worldwide.
We have now generated groundbreaking new tools that are dramatically more efficient and allow targeting more genes per plant with much higher CAS cleavage efficiency. Expect exciting publications soon!
More recently, we showed its affcitveness in tomato (Nature Comm 2025).
Plant growth, development, and response to the environment are mediated by a group of small signaling molecules named hormones. Plants regulate hormone response pathways at multiple levels, including biosynthesis, metabolism, perception, and signaling. In addition, plants exhibit the unique ability to spatially control hormone distribution. Our group identified multiple transporters for key plant hormones.
Auxin transporter (Zhang et al., 2018, Nature Comm, Hu et al., 2021 Nature Comm, Chen al., 2023 EMBO Reports)
Cytokinin (Hu et al., 2023 Nature Plants, Hu et al., 2023 New Phytologist),
Gibberellin (Tal et al., 2016 Nature Comm, Binenbaum et al., 2023 Nature Plants).
ABA (Zhang et al., 2021 Science Advances).
.jpg)
Plant Hormone Transport

Gibberellin and abscisic acid transporters facilitate endodermal suberin formation in Arabidopsis
Distribution patterns and finely tuned concentration gradients of plant hormones govern plant growth and development. Gibberellin (GA) is a plant hormone that regulates key processes in plants, many of them are of significant agricultural importance, such as seed germination, root and shoot elongation, flowering, and fruit patterning. Although studies have demonstrated that GA movement is essential for multiple developmental aspects, how GAs are transported throughout the plant and where exactly does it accumulates remain largely unknown (Binenbaum et al., 2018, Trends in Plant Science.)
Our lab has identified NPF3 is a GA and ABA transporter. NPF3 knockout plants are defective in the accumulation of both GA3-Fl and GA4-Fl in the root endodermis elongation zone. Transport assays in Xenopus oocytes demonstrated that NPF3 can import GAs across membranes (Tal et al., 2016, Nature Comm).
We recently reported that a set of three redundant transporters regulate gibberellin precursors from the shoot to the root to regulate suberin formation. Not only is this the first report on the genetic components required for shoot-to-root gibberellin transport, and that identified the first tonoplast gibberellin transporters in plants, but we were also able to report for the first time on the requirement of gibberellin in suberin regulation in the root endodermis (Binenbaum et al., 2023 Nature Plants)

ABA homeostasis and long-distance translocation
Abscisic acid (ABA) is a plant hormone that regulates growth and responses to the changing environment. For example, seed dormancy, germination, drought tolerance, stomatal closure and lateral root emergence are modulated by this important hormone under normal conditions and in response to stimuli.
We exploited a genome-scale amiRNA screen targeting the Arabidopsis transportome to identify novel ABCG ABA transporters. ABCG17 and ABCG18 are localized to the plasma membrane and import ABA. ABCG17- and ABCG18-knockdown lines displayed enhanced ABA accumulation, induced ABA response, and reduced stomatal aperture size, conductance, and transpiration with increased water-use efficiency compared to WT. In addition, seedlings with double-loss-of-function of ABCG17 and ABCG18 showed enhanced long-distance ABA translocation from the shoot to the root, which led to lateral root outgrowth inhibition. ABCG17 and ABCG18 are primarily expressed in the shoot mesophyll and stem cortex cells, where they drive ABA import, allowing ABA-GE formation. In turn, these ABA-GE sinks prevent active ABA accumulation in guard cells and limit ABA long-distance travel to lateral root formation sites. Under abiotic stress conditions, ABCG17 and ABCG18 are transcriptionally repressed, promoting active ABA movement, accumulation, and response (Zhang et al., 2021, Sciense Advances)

Cytokinin activity–transport and homeostasis at the whole plant, cell, and subcellular levels
Another example of our recent mechanistic work is the identification of three previously unstudied genes, PUP7, PUP21, and PUP8, which belong to the same phylogenetic clade and encode cytokinin (CK) transporters with partially overlapping functions (Hu et al., 2023, New Phytologist). Transport activity and subcellular localization assays showed that PUP7 and PUP21 are CK transporters localized in the tonoplast and that PUP8 is localized to the plasma membrane. Together, these transporters work in concert to establish the spatial pattern of CK signaling within the shoot apical meristem, thereby redundantly controlling meristem size, phyllotaxis, and plant growth (Hu et al., 2023, Nature Plants)

Auxin transport
Auxin (IAA) is a small organic molecule that coordinates many of the key processes in plant development and adaptive growth. Plants regulate the auxin response pathways at multiple levels including biosynthesis, metabolism, and perception. In addition, plants exhibit the unique ability to spatially regulate hormone distribution. This ability is illustrated most clearly in the case of auxin (IAA). The combined activity of auxin influx and efflux carrier proteins generates auxin maxima and local gradients that inform developmental patterning. The regulation of the cellular localization of PIN-FORMED (PIN) efflux transporters determines the direction of auxin flow from one cell to another. Using a transportome, multi-targeted forward-genetic screen artificial microRNAs (amiRNAs) approach we reviled ABCB6 and ABCB20 genes as auxin transport required for shoot growth (Zhang et al., 2018, Nature Comm) and ABCB15-22 as auxin transport in the outer tissues of the root meristem to regulate lateral root spacing (Chen et al., 2023, EMBO Reports).

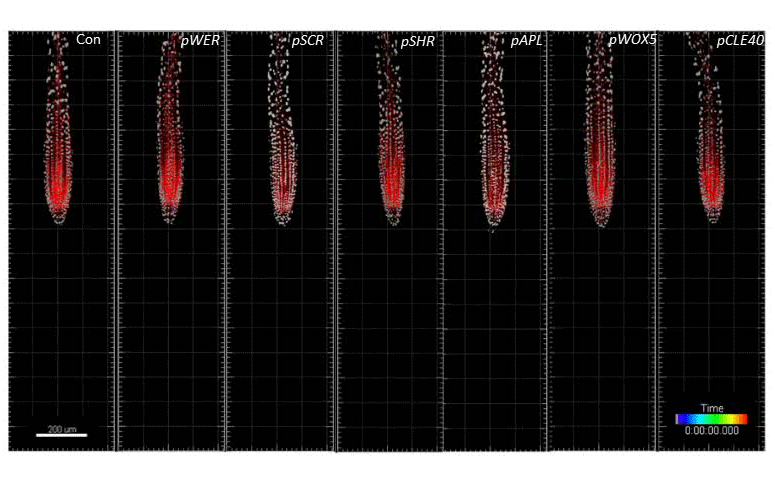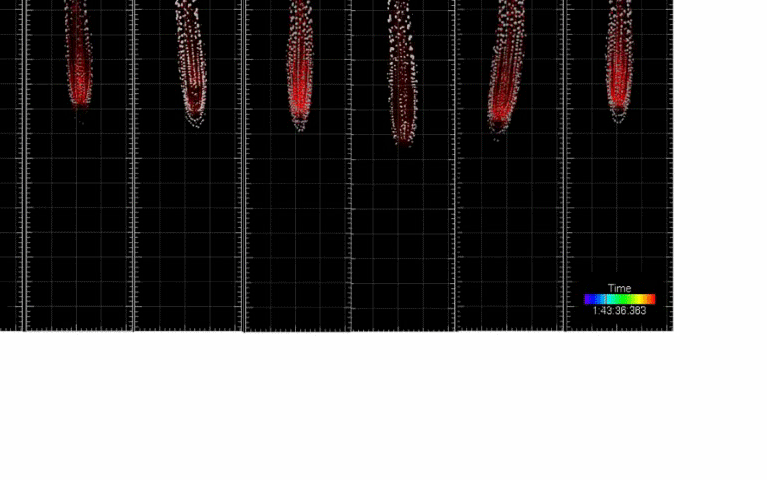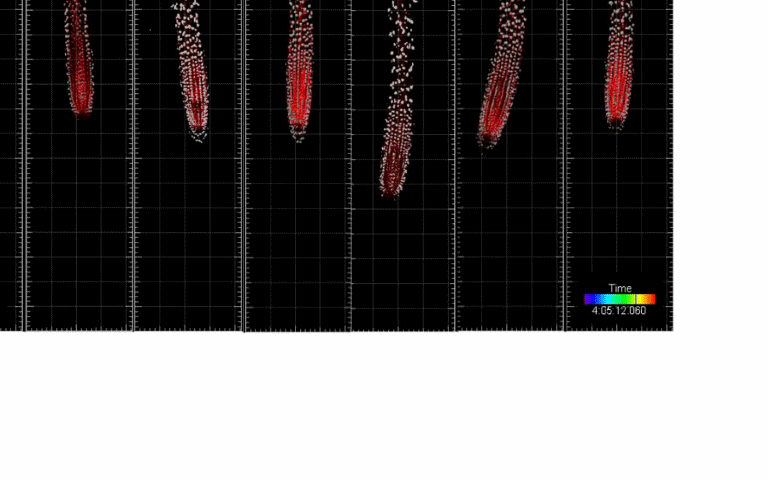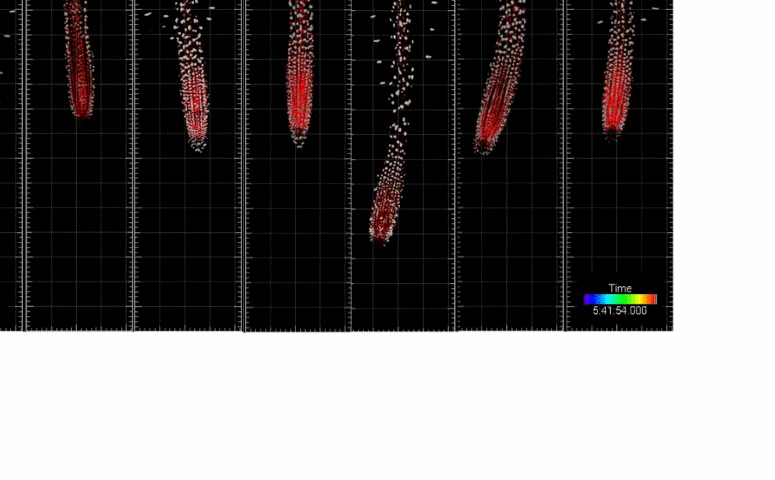
Single-cell kinetics of hormone transport and activity
We developed toolboxes to study hormone-mediated root response and movement that features: (i) the ability to control the hormone synthesis with high spatio-temporal resolution and (ii) single-cell nucleus tracking and morphokinetic analysis infrastructure. Integration of these two features enables cutting-edge analysis of root development at a single-cell resolution based on morphokinetic parameters under normal growth conditions and during cell-type-specific induction of the hormone biosynthesis. Using this approach we mapped directional auxin flow in the root and refined the contributions of key players in this process. In addition, single-cell tracking allowed us to determine the quantitative kinetics of Arabidopsis root meristem skewing (Hu et al., 2021, Nature Comm)